Like accents over letters can be quintessential to correctly interpret a text, the modifications of nucleic acid can have important roles in deciphering the genetic information. While the biological function of the DNA epigenetic modifications, i.e. those that do not affect the genetic sequence has been extensively studied for decades, the similar, so called epitranscriptomic modifications of RNA got started to be deciphered only recently. Researchers of the ELKH Research Centre for Natural Sciences, the Eötvös Loránd University, the Budapest University of Technology and Economics and the Semmelweis University identified the molecular underpinnings of one such RNA transformation mechanism. They also showed that while the impairment of this mechanism can lead to genetic diseases, its deeper understanding can also lead to potentially ground-breaking novel therapeutic applications.
The isomerization of uridine to pseudouridine is the most abundant posttranscriptional modification of RNA and this reaction is catalyzed by a huge protein-RNA complex named box H/ACA pseudouridine synthase. Mutations appearing in its protein components are linked to serious illnesses like bone marrow failure, cancer or nephrotic syndrome.
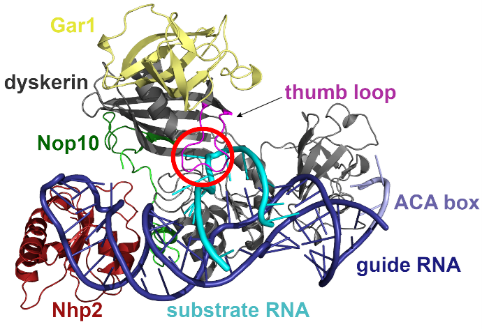
The researchers built a model of the pseudouridine synthase complex and performed large scale computational simulations to reveal the atomistic details of the uridine to pseudouridine transformation. They found that the RNA to be modified is bound to the box H/ACA pseudouridine synthase in a distorted form, which is well prepared for the subsequent transformation of the uridine to pseudouridine. They also showed that mutant variants of the enzyme do not distort the critical uridine, in line with the inactivity of these variants.
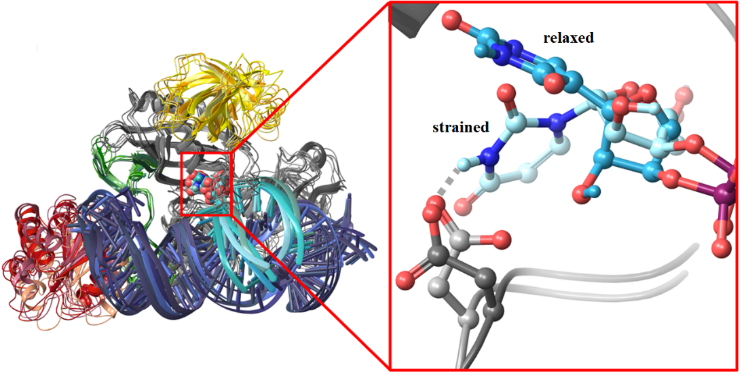
Researchers pointed out that the identified transformation mechanism is able to convert uridine in any RNA that fits well to the guide RNA of the box H/ACA pseudouridine synthase, and the structure of the guide RNA can vary to a large extent. Thus, engineered guide RNAs that can associate with the pseudouridine synthase complex may be applied to facilitate the endogenous pseudouridylation of nearly any substrate. This suggests the opportunity of programmable RNA edition for a wide range of RNAs and therapeutics for gene lesion related diseases.
Project no. K111862, K116305 and FK124230 have been implemented with the support provided by the Ministry of Innovation and Technology of Hungary from the National Research, Development and Innovation Fund, financed under the OTKA funding scheme. The KIFÜ-NIIF Institute granted computational time on the Hungarian HPC Infrastructure.
Dóra Judit Kiss, Julianna Oláh, Gergely Tóth, Máté Varga, András Stirling, Dóra K. Menyhárd, György G. Ferenczy
The structure-derived mechanism of box H/ACA pseudouridine synthase offers a plausible paradigm for programmable RNA editing

