Introduction
The non-coding genome encompasses a group of genes that are transcribed, but do not possess protein-coding capability. For a long time, this part of the genome was considered “junk DNA,” and no physiological significance was attributed to it. However, this view gradually changed, first with the discovery of microRNAs and later with the recognition of long non-coding RNAs, known as lncRNAs. The Non-Coding Genome research group primarily focuses on the study of long non-coding RNAs, that are represented by about 58,000 transcripts in the human genome. These RNAs exhibit altered expression in various pathological conditions, linking them to the development and progression of cancer, neurodegenerative diseases, psychological disorders, and autoimmune diseases, among others. The main goal of the research group is to uncover the physiological effects and molecular mechanisms of lncRNAs.
Ongoing research
The roles of long non-coding RNAs in cancer
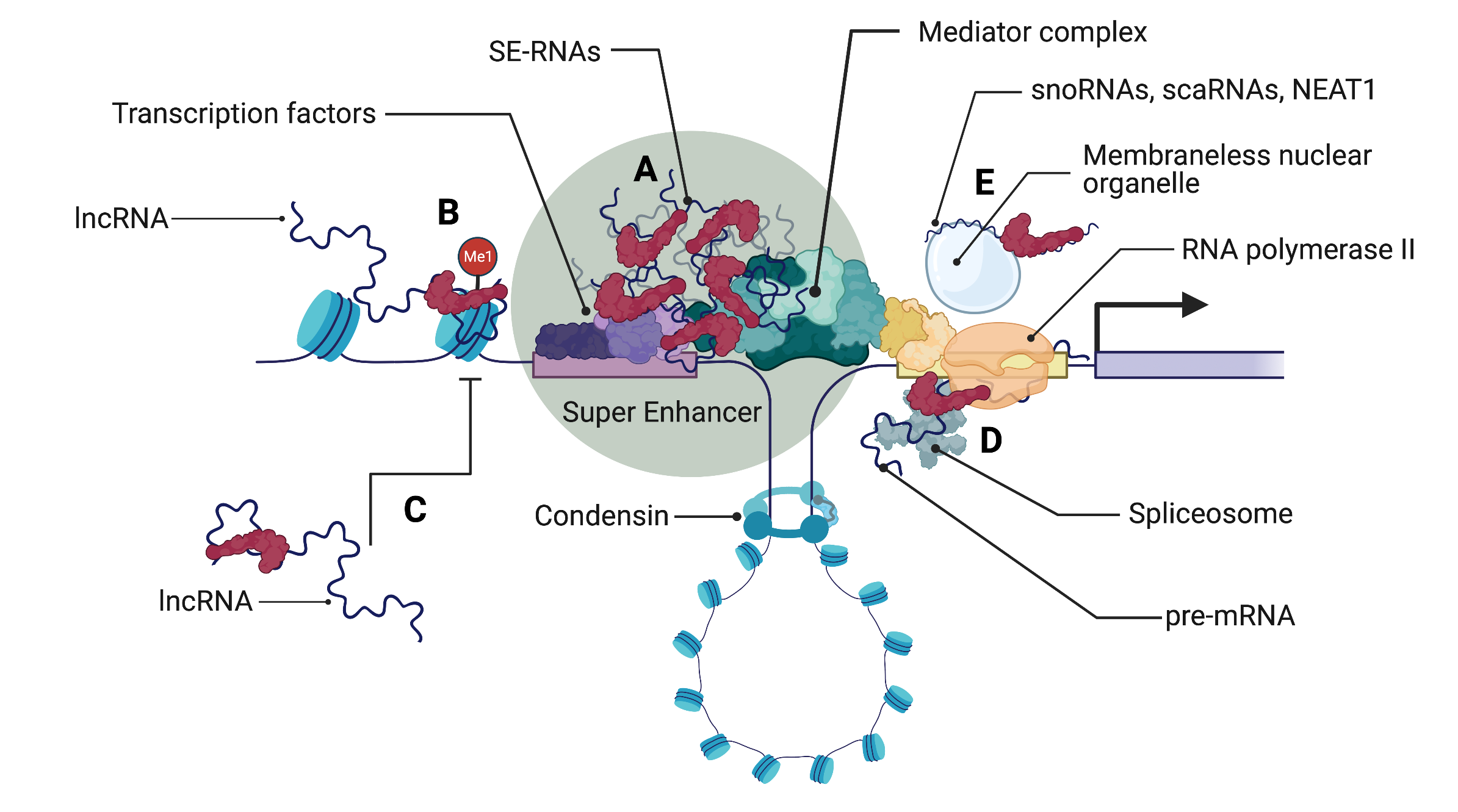
Long non-coding RNAs (lncRNAs) play a regulatory role in numerous cellular processes, often manifested through molecular interactions. Due to their large size and flexibility, they can bind to a considerable number of partner molecules simultaneously, exerting robust effects even with low expression levels. Their significance is well-known in various cellular functions, including the regulation of the cell cycle, DNA repair, stress responses, gene expression, and RNA processing. Thus, it is not surprising that their abnormal expression is associated with the development and progression of several cancers, making them not only potential drug targets but also abundant sources of biomarker candidates.
However, it is important to note that these RNAs rarely function independently, and their roles and effects are often redundant, so achieving significant therapeutic impact through the regulation of a single lncRNA is unlikely. Therefore, in the Non-Coding Genome Research Group, we aim to investigate these lncRNAs using a network-based approach, considering both their protein and various RNA partners. With this method, we can identify significant hub lncRNAs, whose collective targeting may result in substantial therapeutic effects and can be utilized as robust diagnostic and prognostic markers.
RNA binding of Histone lysine methyltransferases
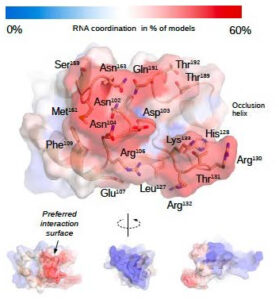
The post-translational modification of histone proteins is a key element in the regulation of the genetic program. Among the numerous histone-modifying enzymes, the focus of the Non-Coding Genome Research Group is on histone lysine methyltransferases (HKMTs), specifically their various RNA interactions. Several HKMTs are known to have significant RNA-binding capabilities, with these interactions regulating the enzymes’ canonical activities, while non-canonical functions are also associated with HKMT-RNA interactions.
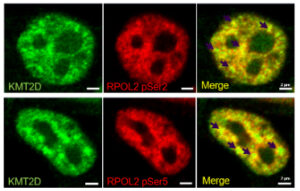
The research group was the first to describe the RNA partners of two HKMTs, KMT2D and KMT2F (SETD1A), as well as their primary RNA-binding regions. In vitro experiments confirmed the RNA binding for numerous partners. The next stage of the research focuses on the role of these two proteins in RNA processing and the function of specific interactions with selected lncRNAs. The involvement of HKMTs in RNA processing expands their physiological role with a new function, which is supported by the co-transcriptional RNA binding, the presence of splicing-related RNAs in their interactome, and their intracellular localization.
In the case of KMT2D, special attention is given to the RNA-binding region, as point mutations in this protein segment were identified as the underlying cause of a developmental disorder identified in 2020. Preliminary results indicate that mutations affect the structure and stability of the RNA-binding region, its phase transition capability, and the specificity of RNA recognition. The research group collaborates with the researchers who first described the disease to uncover the molecular connections between mutations and the development of the disorder.
Efficient characterization and prediction of liquid-liquid phase separation systems
One of the significant scientific discoveries in recent years is the liquid-liquid phase separation (LLPS) of proteins, which has been confirmed to be a fundamental aspect of most cellular processes. LLPS plays a central role in the development of neurodegenerative diseases, but it is also crucial in cancer-related processes, cellular stress responses, and the regulation of the cell cycle. Therefore, it is extremely important to understand the molecular and biophysical mechanisms that enable and regulate LLPS. As a result, the bioinformatic characterization of proteins capable of LLPS is of paramount importance.
Several attempts have been made to identify these proteins based on their sequences, but due to the extraordinary complexity of the process, these efforts have remained limited in efficiency. The Non-Coding Genome Research Group approaches the problem with a novel method by collectively characterizing multiple components involved in LLPS systems. The group is working on the development of a truly effective LLPS prediction algorithm that accurately describes physiological phenomena.
Databases
Database of Proteins Driving Liquid-liquid Phase Separation (PhasePro): https://phasepro.elte.hu
Database of Disordered Binding Sites (DIBS): https://dibs.enzim.ttk.mta.hu
Protein Ensemble Database (PED): https://proteinensemble.org
Collaborations
International collaborations
The University of Manchester, UK (https://research.manchester.ac.uk/en/persons/siddharth.banka)
Thomas Jefferson University, USA; Sidney Kimmel Cancer Center, USA (https://www.pcarmrc.org/pestell-1)
University of Novi Sad, Serbia (https://www.dbe.uns.ac.rs/vanredni-profesori/zeljko-popovic/)
European Molecular Biology Laboratory Heidelberg, Germany
Collaborations with HUN-REN Institutes
HUN-REN Wigner Research Centre for Physics, Hungary
National Collaborations
Budapest University Of Technology And Economics, Hungary
Eötvös Loránd University (http://abnmr.elte.hu), Hungary
Industrial Partners
BBS Nanotechnology Ltd., Hungary
PhD Students
Szalainé Ágoston Bianka (year of graduation: 2013)
Murvai Nikoletta (year of graduation: 2021)
Harem Sabr Muhamad Amin (predicted year of graduation: 2023)
Mevan Jacksi Fahmi (predicted year of graduation: 2023)
Mustafa Abdulkareem (predicted year of graduation: 2027)
Important publications
1. Amin, H.M.; Abukhairan, R.; Szabo, B.; Jacksi, M.; Varady, Gy.; Lozsa, R.; Schad, E.; Tantos, A. (2023) KMT2D preferentially binds mRNAs of the genes it regulates, suggesting a role in RNA processing. PROTEIN SCIENCE 33 : 1 Paper: e4847
2. Amin, H.M.; Szabo, B.; Abukhairan, R.; Zeke, A.; Kardos, J.; Schad, E.; Tantos, A. (2023) In Vivo and In Vitro Characterization of the RNA Binding Capacity of SETD1A (KMT2F). INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES 24 : 22 Paper: 16032
3. Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Zeke, A.; Lazar, T.; Glavina, J.; Nagy-Kanta, E.; Donagh, J.-M.; Kalman, Zs.E.; Pascarelli, S. et al. (2023) ELM—the Eukaryotic Linear Motif resource—2024 update. NUCLEIC ACIDS RESEARCH 2023 p. 1 Paper: gkad1058
4. Mészáros, B.; Hatos, A.; Palopoli, N.; Quaglia, F.; Salladini, E.; Van Roey, K.; Arthanari, H.; Dosztányi, Zs.; Felli, I.C.; Fischer, P.D. et al. (2023) Minimum information guidelines for experiments structurally characterizing intrinsically disordered protein regions. NATURE METHODS 20 pp. 1291-1303. , 13 p.
5. Szabó, Cs.L.; Szabó, B.; Sebák, F.; Bermel, W.; Tantos, A.; Bodor, A. (2022) The Disordered EZH2 Loop: Atomic Level Characterization by 1HN- and 1Hα-Detected NMR Approaches, Interaction with the Long Noncoding HOTAIR RNA. INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES 23 : 11 Paper: 6150
6. Zeke, A. ; Schád, É. ; Horváth, T. ; Abukhairan, R. ; Szabó, B. ; Tantos, A. (2022) Deep structural insights into RNA-binding disordered protein regions. WILEY INTERDISCIPLINARY REVIEWS-RNA 13 : 5 Paper: e1714
7. Pancsa, R.; Fichó, E.; Molnár, D.; Surányi, É.V.; Trombitás, T.; Füzesi, D.; Lóczi, H.; Szijjártó, P.; Hirmondó, R.; Szabó, J.E. et al. (2022) dNTPpoolDB: a manually curated database of experimentally determined dNTP pools and pool changes in biological samples. NUCLEIC ACIDS RESEARCH 50 : D1 pp. 1508-1514. , 7 p.
8. Necci, M.; Piovesan, D.; Hoque, M.T.; Walsh, I.; Iqbal, S.; Vendruscolo, M.; Sormanni, P.; Wang, C.; Raimondi, D.; Sharma, R. et al. (2021) Critical assessment of protein intrinsic disorder prediction. NATURE METHODS 18 : 5 pp. 472-481. , 10 p.
9. Korkmazhan, E.; Tompa, P.; Dunn, A.R. (2021) The role of ordered cooperative assembly in biomolecular condensates. NATURE REVIEWS MOLECULAR CELL BIOLOGY 22 : 10 pp. 647-648. , 2 p.
10. Mészáros, B.; Erdős, G.; Szabó, B.; Schád, É.; Tantos, Á.; Abukhairan, R.; Horváth, T.; Murvai, N.; Kovács, O.P.; Kovács, M. et al. (2020) PhaSePro: the database of proteins driving liquid-liquid phase separation. NUCLEIC ACIDS RESEARCH 48 : D1 pp. D360-D367.
Group Leader
Agnes Tantos (Publications)
Members
Peter Tompa, DSc (Publications)
Eva Schad, PhD (Publications)
Beata Nemethne Szabo, MSc (Publications)
Rita Pancsa, PhD (Publications)
Mehvan Jacksi, MSc
Mustafa Abdulkareem. MSc