 Introduction, main research areas
Introduction, main research areas
The Protein Bioinformatics Research Group at the Molecular Institute of Life Sciences, Research Centre of Natural Sciences is headed by Dr. Gabor Tusnady, and was founded in 2019, when the Protein Structure Research Group and the Transmembrane Protein Bioinformatics Research Group were fused.
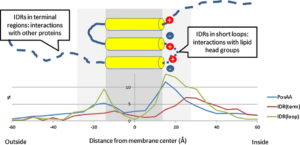
The group’s main aim is to investigate membrane proteins topology and 3D structure by developing state-of-the-art computational techniques, like CCTOP or TMFoldRec algorithms, as well as to establish and maintain databases relating to transmembrane protein structure and topology. The group also investigates the disordered regions in transmembrane and globular proteins and the interactions of transmembrane proteins. Beyond bioinformatics studies, the group is involved in developing a high throughput experimental method for generating topology data of transmembrane proteins.
In addition to these recent achievements in the field of transmembrane proteins, the group was involved in several studies using next generation sequencing (NGS) ranging from genome sequencing of the chicken DT40 bursal lymphoma cell line and Mycoplasma sp. HU2014, through studying mutagenic impact of common cancer cytotoxics and clinical validation of TruSeq Custom Amplicon assay performed on formalin-fixed and paraffin-embedded tumor samples, to the identification of known and unknown plant viral pathogens.
Recent collaborators
- Pediatrics, Harvard Medical School
- National Institute of Oncology
- University of Szeged
- Institute of Organic Chemistry, RCNS
Main equipment
- Supermicro computer cluster (1240 CPU core, 2.5
 Tb RAM, 240 Tb redundant winchester capacity.
Tb RAM, 240 Tb redundant winchester capacity. - Web server (40 CPU core, 160Gb RAM, 12Tb redundant Sata disk, 2x4Tb PCIexpress SSD disk)
- Web server (48 CPU core, 390Gb RAM, 33Tb redundant Sata disk)
- 60Tb QNAP NAS server
PhD students in the last 10 years
Best 10 publications in the last 5 years
- Dobson, L, Gerdán, C, Tusnády, S, Szekeres, LI, Kuffa, KR, Langó, T, Zeke, A and Tusnády, GE (2024) UniTmp: unified resources for transmembrane proteins. Nucleic Acids Res 52: D572-D578.
- Dobson, L, Szekeres, LI, Gerdán, C, Langó, T, Zeke, A and Tusnády, GE (2023) TmAlphaFold database: membrane localization and evaluation of AlphaFold2 predicted alpha-helical transmembrane protein structures. Nucleic Acids Res 51: D517-D522.
- Jambrich, MA, Tusnady, GE and Dobson, L (2023) How AlphaFold2 shaped the structural coverage of the human transmembrane proteome. Sci Rep 13: 20283.
- Zeke, A, Takács, T, Sok, P, Németh, K, Kirsch, K, Egri, P, Póti, ÁL, Bento, I, Tusnády, GE and Reményi, A (2022) Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling. Nat Commun 13: 5439.
- Langó, T, Kuffa, KR, Tóth, G, Turiák, L, Drahos, L and Tusnády, GE (2022) Comprehensive Discovery of the Accessible Primary Amino Group-Containing Segments from Cell Surface Proteins by Fine-Tuning a High-Throughput Biotinylation Method. Int J Mol Sci 24: 273.
- Zeke, A, Dobson, L, Szekeres, LI, Langó, T and Tusnády, GE (2021) PolarProtDb: A Database of Transmembrane and Secreted Proteins showing Apical-Basal Polarity. J Mol Biol 433: 166705.
- Dobson, L, Zeke, A and Tusnády, GE (2021) PolarProtPred: predicting apical and basolateral localization of transmembrane proteins using putative short linear motifs and deep learning. Bioinformatics 37: 4328-4335.
- Dobson, L and Tusnády, GE (2021) MemDis: Predicting Disordered Regions in Transmembrane Proteins. Int. J. Mol. Sci. 22: 12270.
- Langó, T, Pataki, ZG, Turiák, L, Ács, A, Varga, JK, Várady, G, Kucsma, N, Drahos, L and Tusnády, GE (2020) Partial proteolysis improves the identification of the extracellular segments of transmembrane proteins by surface biotinylation. Sci Rep 10: 8880.
- Müller, A, Langó, T, Turiák, L, Ács, A, Várady, G, Kucsma, N, Drahos, L and Tusnády, GE (2019) Covalently modified carboxyl side chains on cell surface leads to a novel method toward topology analysis of transmembrane proteins. Sci Rep 9: 15729.


